Chris West Lab
DNA damage and repair
Research interests
Research interests
DSB repair mechanisms
Non-homologous end-joining
Transgene integration
DNA damage responses
Genome maintenance in germination and seed longevity
DNA repair and recombination mechanisms in plants
We are focussed on understanding how plants repair damage to their DNA and the roles of these mechanisms in crop responses to environmental stress and genome modification in plant biotechnology.
DNA is important for plant growth and the transmission of genetic material between generations but is constantly under attack from environmental factors such as UVB, soil pollutants, and the by-products of cell metabolism including reactive oxygen species. If unrepaired, DNA damage can inhibit growth, threaten cellular survival and cause mutation of the genome. Cells have powerful mechanisms which detect and repair DNA damage, minimising the deleterious consequences to the plant cell.
Our research is focussed on understanding how plants respond and repair to DNA damage, in particular DNA double strand breaks, a highly cytotoxic and mutagenic lesion in which both strands of the chromosome are severed. These repair mechanisms, termed recombination, are important for plant responses to environmental stress but also underpin genome modification in plant biotechnology. Translation of our fundamental research findings has important applications to improvement crop performance under rapidly changing climates and the development of optimised genome editing methodologies in plant biotechnology. Furthermore, our recent work has revealed that DNA repair and response mechanisms are important factors which determine germination performance and seed longevity, major determinants of crop yields.
DSB repair mechanisms
DSBs can be repaired by two mechanisms which are highly conserved throughout eukaryotic evolution: either homology-based repair or simple end-to-end joining. In homologous recombination (HR), a break is re-joined using a sister chromatid, homologue or other identical/near identical sequence as a template for repair synthesis to ensure faithful restoration of the original sequence. In contrast, broken chromosomes can also be repaired by illegitimate recombination (IR) or non-homologous end joining (NHEJ) in which DSBs are simply re-joined, end-to-end, independent of DNA sequence.
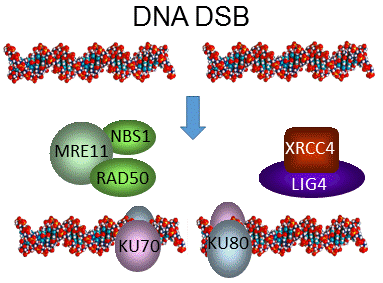
Non-homologous end-joining
The NHEJ pathway of DSB repair can be summarised in three stages: i) recognition of DNA damage; ii) processing and juxtaposition of the DNA ends and iii) ligation (re-joining). These processes involve the formation of protein complexes which catalyse these reactions. In the early stages of DNA repair a complex of KU70 and KU80 bind to DNA ends at sites of DSBs in the DNA. The DNA ends may then be processed by the MRE11-RAD50-NBS1 (MRN) complex to make them suitable substrates for joining by the DNA Ligase IV (AtLIG4)/ XRCC4 complex. In addition to this conserved pathway, alternative pathways are active in plants. Current efforts are identifying mechanisms and factors involved, in collaboration with other groups in the EU.
Transgene integration
DNA introduced into a cell (a transgene) is often stably incorporated into the cell’s own DNA (genome), in a process that uses the host’s DNA repair / recombination pathways. Transgene integration is a very important tool for scientific research and for biotechnology, and understanding the mechanisms of integration provide insight into molecular basis of this technology. In collaboration with groups in the US, we have investigated the roles of DSB repair pathways in transgene integration..
DNA damage response
The DNA damage response (DDR) is a complex cellular network which integrates genome surveillance and signalling with downstream responses, activating DNA repair factors, cell cycle progression and pathways of programmed cell death (PCD). The eukaryotic cellular response to DNA damage is orchestrated by the phosphoinositide-3-kinase-related protein kinases (PIKKs) ATAXIA TELANGIECTASIA MUTATED (ATM) and ATM AND RAD3-RELATED (ATR). In response to genotoxic stresses, these checkpoint kinases activate DNA repair factors, delay or halt cell cycle progression, and promote endocycles or programmed cell death (PCD). ATM principally responds to DSBs whilst ATR principally in activated in response to DNA replication stress. Current projects focus on elucidating the dynamics of post-translational signalling in the plant DDR using high throughput approaches (phosphoproteomics, transcriptomics, yeast two-hybrid). This has proven a powerful way to identify novel factors important to genome stability in plants, and has identified histone modifications associated with the response to DNA damage.
Seeds: the importance of safeguarding the genome in germination
Successful germination is fundamental for agriculture and plant survival in the natural environment. Embryo viability is effectively prolonged in the dry quiescent state of the desiccation tolerant seed, which are the predominant seeds of crop species and plants that grow in temperate latitudes. However, extended periods in this desiccated state result in compromised germination and associated losses in crop yield. We have identified important roles for DSB repair and response mechanisms in the seed stage of the plant lifecycle, which function to safeguard the germinating embryo from extensive genome damage incurred in the desiccated state. DSB repair becomes particularly important in germination and seed storage under adverse stress conditions.

